Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 1192-1-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1836 | 1.034e-02 | 1.236e-02 | 1.016e-01 |
|---|
| IPC-NIBC-129 | 0.2764 | 4.732e-02 | 6.003e-02 | 4.360e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2467 | 1.782e-02 | 2.588e-02 | 3.659e-01 |
|---|
| Loi_GPL570 | 0.2158 | 4.277e-02 | 4.244e-02 | 2.268e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1568 | 9.200e-05 | 4.600e-04 | 1.913e-03 |
|---|
| Parker_GPL1390 | 0.2623 | 1.234e-01 | 1.252e-01 | 5.967e-01 |
|---|
| Parker_GPL887 | 0.0601 | 8.010e-01 | 8.199e-01 | 8.551e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3967 | 3.484e-01 | 3.093e-01 | 9.023e-01 |
|---|
| Schmidt | 0.2070 | 1.241e-02 | 1.756e-02 | 1.542e-01 |
|---|
| Sotiriou | 0.3020 | 2.091e-03 | 5.770e-04 | 5.475e-02 |
|---|
| Wang | 0.2160 | 1.165e-01 | 8.218e-02 | 4.032e-01 |
|---|
| Zhang | 0.1447 | 6.224e-02 | 3.712e-02 | 1.562e-01 |
|---|
| Zhou | 0.4236 | 4.312e-02 | 4.404e-02 | 2.765e-01 |
|---|
Expression data for subnetwork 1192-1-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
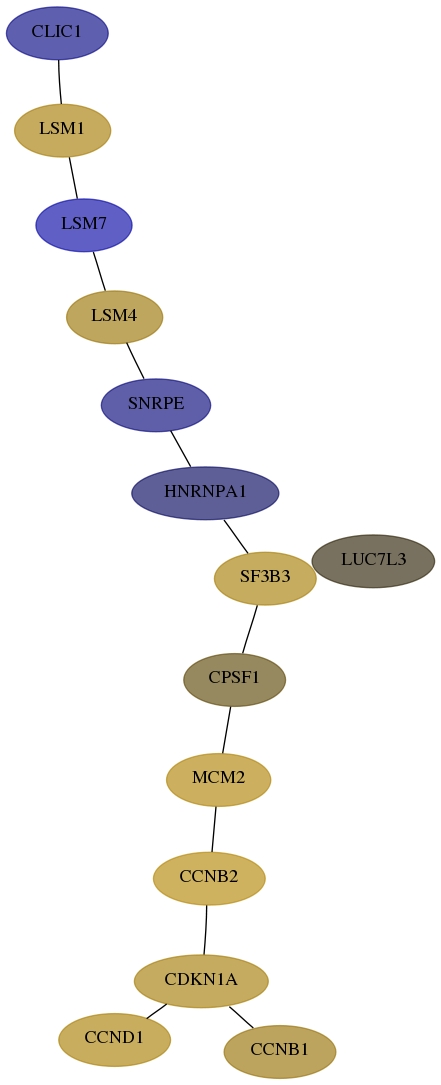
Score for each gene in subnetwork 1192-1-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| snrpe |   | 1 | 164 | 370 | 370 | -0.041 | 0.274 | 0.118 | 0.012 | 0.114 | 0.100 | 0.077 | 0.134 | 0.124 | 0.105 | 0.094 | 0.028 | 0.176 |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| clic1 |   | 1 | 164 | 370 | 370 | -0.053 | 0.208 | 0.089 | 0.010 | 0.123 | 0.236 | 0.400 | 0.229 | 0.084 | 0.094 | 0.055 | -0.037 | 0.159 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| cpsf1 |   | 3 | 79 | 282 | 264 | 0.020 | -0.019 | 0.031 | 0.075 | 0.102 | 0.120 | 0.204 | 0.157 | -0.009 | 0.093 | 0.095 | 0.089 | 0.102 |
|---|
| mcm2 |   | 4 | 60 | 216 | 192 | 0.151 | 0.254 | 0.188 | 0.128 | 0.154 | 0.220 | -0.034 | 0.267 | 0.224 | 0.200 | 0.101 | 0.012 | 0.255 |
|---|
| ccnb1 |   | 36 | 7 | 78 | 59 | 0.082 | 0.246 | 0.212 | 0.224 | 0.256 | 0.099 | 0.126 | 0.351 | 0.129 | 0.249 | 0.152 | 0.167 | 0.398 |
|---|
| lsm7 |   | 1 | 164 | 370 | 370 | -0.113 | 0.189 | 0.116 | 0.137 | 0.045 | 0.132 | 0.126 | 0.283 | 0.094 | -0.037 | -0.022 | 0.053 | 0.166 |
|---|
| luc7l3 |   | 7 | 38 | 1 | 5 | 0.006 | 0.111 | -0.046 | -0.016 | 0.121 | 0.071 | -0.318 | -0.084 | 0.159 | 0.098 | 0.155 | 0.067 | 0.065 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| lsm1 |   | 1 | 164 | 370 | 370 | 0.118 | 0.094 | 0.025 | 0.167 | 0.118 | -0.014 | 0.083 | 0.229 | 0.012 | 0.034 | 0.089 | 0.149 | 0.172 |
|---|
| hnrnpa1 |   | 4 | 60 | 1 | 9 | -0.021 | 0.209 | 0.088 | -0.020 | 0.087 | 0.131 | undef | 0.082 | 0.036 | 0.089 | 0.077 | 0.083 | 0.195 |
|---|
| sf3b3 |   | 7 | 38 | 1 | 5 | 0.119 | 0.241 | 0.143 | -0.019 | 0.082 | 0.222 | 0.007 | 0.271 | 0.168 | 0.060 | 0.017 | -0.077 | 0.372 |
|---|
| lsm4 |   | 1 | 164 | 370 | 370 | 0.089 | 0.169 | 0.110 | 0.197 | 0.156 | 0.060 | 0.195 | 0.166 | 0.052 | 0.126 | 0.032 | 0.072 | 0.324 |
|---|
GO Enrichment output for subnetwork 1192-1-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.662E-07 | 3.823E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 2.373E-07 | 2.729E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.26E-07 | 2.499E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 3.26E-07 | 1.874E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 4.342E-07 | 1.998E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 9.986E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.479E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.418E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.062E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 6.693E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 7.241E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 7.178E-08 | 1.754E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 1.314E-07 | 1.606E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 1.708E-07 | 1.391E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 2.172E-07 | 1.327E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 4.051E-07 | 1.979E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.364E-06 | 5.552E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.614E-06 | 5.634E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.939E-06 | 5.921E-04 |
|---|
| spliceosome assembly | GO:0000245 |  | 3.529E-06 | 9.579E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.889E-06 | 2.172E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.29E-05 | 2.864E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.065E-07 | 2.563E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 1.95E-07 | 2.346E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 2.534E-07 | 2.032E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.222E-07 | 1.938E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 6.008E-07 | 2.891E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.021E-06 | 8.105E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.39E-06 | 8.216E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.567E-06 | 7.72E-04 |
|---|
| interphase | GO:0051325 |  | 2.707E-06 | 7.236E-04 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.338E-06 | 1.044E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 2.877E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 7.178E-08 | 1.754E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 1.314E-07 | 1.606E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 1.708E-07 | 1.391E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 2.172E-07 | 1.327E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 4.051E-07 | 1.979E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.364E-06 | 5.552E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.614E-06 | 5.634E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 1.939E-06 | 5.921E-04 |
|---|
| spliceosome assembly | GO:0000245 |  | 3.529E-06 | 9.579E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 8.889E-06 | 2.172E-03 |
|---|
| ribonucleoprotein complex assembly | GO:0022618 |  | 1.29E-05 | 2.864E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.065E-07 | 2.563E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 1.95E-07 | 2.346E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 2.534E-07 | 2.032E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.222E-07 | 1.938E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 6.008E-07 | 2.891E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.021E-06 | 8.105E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.39E-06 | 8.216E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.567E-06 | 7.72E-04 |
|---|
| interphase | GO:0051325 |  | 2.707E-06 | 7.236E-04 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.338E-06 | 1.044E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 2.877E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 6.378E-09 | 2.278E-05 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.814E-08 | 6.809E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.122E-07 | 1.335E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.608E-07 | 4.113E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.549E-07 | 6.106E-04 |
|---|
| RNA splicing factor activity. transesterification mechanism | GO:0031202 |  | 1.366E-06 | 8.131E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.012E-05 | 5.163E-03 |
|---|
| response to UV | GO:0009411 |  | 1.226E-05 | 5.474E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.255E-05 | 4.979E-03 |
|---|
| superoxide-generating NADPH oxidase activity | GO:0016175 |  | 1.255E-05 | 4.481E-03 |
|---|
| nuclear origin of replication recognition complex | GO:0005664 |  | 1.255E-05 | 4.074E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| small nuclear ribonucleoprotein complex | GO:0030532 |  | 1.474E-09 | 5.377E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.462E-08 | 2.667E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 9.481E-08 | 1.153E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.457E-07 | 1.329E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.258E-07 | 1.647E-04 |
|---|
| interphase | GO:0051325 |  | 2.492E-07 | 1.515E-04 |
|---|
| RNA splicing factor activity. transesterification mechanism | GO:0031202 |  | 5.251E-07 | 2.736E-04 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.177E-06 | 1.449E-03 |
|---|
| response to UV | GO:0009411 |  | 3.65E-06 | 1.48E-03 |
|---|
| mRNA binding | GO:0003729 |  | 4.168E-06 | 1.52E-03 |
|---|
| origin recognition complex | GO:0000808 |  | 4.447E-06 | 1.475E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.065E-07 | 2.563E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 1.95E-07 | 2.346E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 2.534E-07 | 2.032E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.222E-07 | 1.938E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 6.008E-07 | 2.891E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.021E-06 | 8.105E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.39E-06 | 8.216E-04 |
|---|
| spliceosomal snRNP biogenesis | GO:0000387 |  | 2.567E-06 | 7.72E-04 |
|---|
| interphase | GO:0051325 |  | 2.707E-06 | 7.236E-04 |
|---|
| spliceosome assembly | GO:0000245 |  | 4.338E-06 | 1.044E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.315E-05 | 2.877E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.662E-07 | 3.823E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 2.373E-07 | 2.729E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.26E-07 | 2.499E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 3.26E-07 | 1.874E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 4.342E-07 | 1.998E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 9.986E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.479E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.418E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.062E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 6.693E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 7.241E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.662E-07 | 3.823E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 2.373E-07 | 2.729E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.26E-07 | 2.499E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 3.26E-07 | 1.874E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 4.342E-07 | 1.998E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 9.986E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.479E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.418E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.062E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 6.693E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 7.241E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.662E-07 | 3.823E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 2.373E-07 | 2.729E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.26E-07 | 2.499E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 3.26E-07 | 1.874E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 4.342E-07 | 1.998E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 9.986E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.479E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.418E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.062E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 6.693E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 7.241E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| mRNA cleavage | GO:0006379 |  | 1.662E-07 | 3.823E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 2.373E-07 | 2.729E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 3.26E-07 | 2.499E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 3.26E-07 | 1.874E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 4.342E-07 | 1.998E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.605E-06 | 9.986E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 4.5E-06 | 1.479E-03 |
|---|
| interphase | GO:0051325 |  | 4.933E-06 | 1.418E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 2.372E-05 | 6.062E-03 |
|---|
| response to UV | GO:0009411 |  | 2.91E-05 | 6.693E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.463E-05 | 7.241E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| RNA export from nucleus | GO:0006405 |  | 1.338E-07 | 3.174E-04 |
|---|
| mRNA cleavage | GO:0006379 |  | 1.613E-07 | 1.913E-04 |
|---|
| mRNA polyadenylation | GO:0006378 |  | 2.952E-07 | 2.335E-04 |
|---|
| RNA polyadenylation | GO:0043631 |  | 4.877E-07 | 2.893E-04 |
|---|
| mRNA 3'-end processing | GO:0031124 |  | 4.877E-07 | 2.315E-04 |
|---|
| nuclear export | GO:0051168 |  | 5.154E-07 | 2.039E-04 |
|---|
| RNA 3'-end processing | GO:0031123 |  | 7.492E-07 | 2.54E-04 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.773E-06 | 5.258E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 2.839E-06 | 7.484E-04 |
|---|
| interphase | GO:0051325 |  | 3.109E-06 | 7.377E-04 |
|---|
| nuclear import | GO:0051170 |  | 3.251E-06 | 7.013E-04 |
|---|



