Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10985-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2050 | 3.727e-03 | 4.416e-03 | 4.631e-02 |
|---|
| IPC-NIBC-129 | 0.2227 | 6.308e-02 | 8.140e-02 | 5.241e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2959 | 5.318e-03 | 8.723e-03 | 1.774e-01 |
|---|
| Loi_GPL570 | 0.2724 | 8.894e-02 | 8.840e-02 | 3.827e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1594 | 2.000e-06 | 1.400e-05 | 5.000e-06 |
|---|
| Parker_GPL1390 | 0.2413 | 9.169e-02 | 9.070e-02 | 5.184e-01 |
|---|
| Parker_GPL887 | 0.1443 | 5.024e-01 | 5.360e-01 | 5.381e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3871 | 2.840e-01 | 2.446e-01 | 8.505e-01 |
|---|
| Schmidt | 0.2641 | 5.734e-03 | 8.387e-03 | 8.409e-02 |
|---|
| Sotiriou | 0.3490 | 1.657e-03 | 4.300e-04 | 4.613e-02 |
|---|
| Wang | 0.1737 | 1.118e-01 | 7.832e-02 | 3.928e-01 |
|---|
| Zhang | 0.1664 | 1.145e-01 | 7.706e-02 | 2.708e-01 |
|---|
| Zhou | 0.4376 | 4.730e-02 | 4.877e-02 | 3.014e-01 |
|---|
Expression data for subnetwork 10985-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
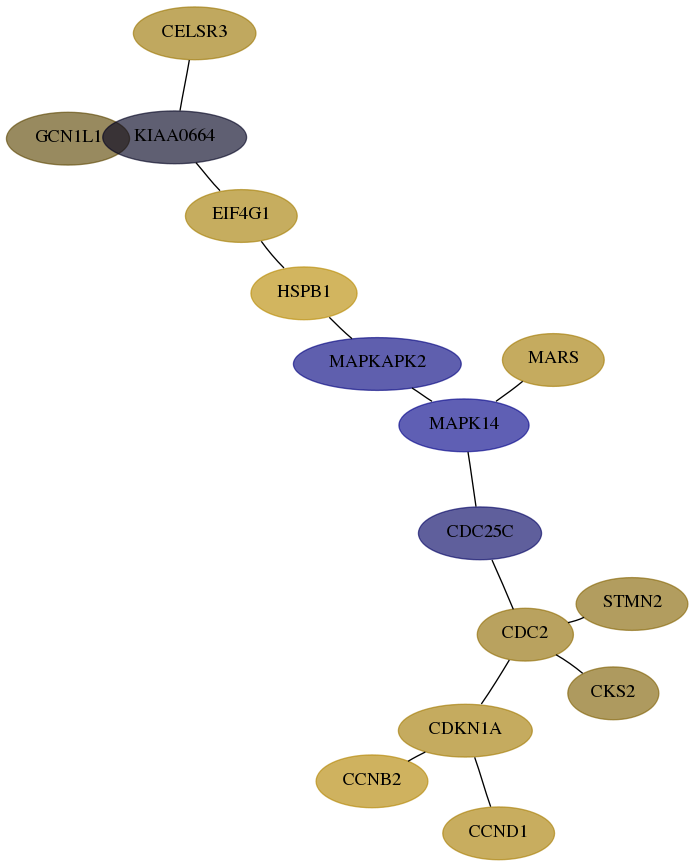
Score for each gene in subnetwork 10985-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| ccnb2 |   | 16 | 13 | 153 | 125 | 0.164 | 0.176 | 0.204 | 0.123 | 0.172 | 0.178 | 0.090 | 0.366 | 0.240 | 0.169 | 0.144 | 0.211 | 0.463 |
|---|
| kiaa0664 |   | 1 | 164 | 153 | 168 | -0.004 | 0.082 | -0.002 | 0.038 | 0.038 | 0.091 | 0.125 | 0.141 | 0.107 | 0.005 | 0.016 | 0.140 | 0.095 |
|---|
| stmn2 |   | 29 | 8 | 1 | 3 | 0.057 | 0.106 | 0.005 | 0.052 | 0.110 | 0.071 | -0.099 | 0.006 | 0.034 | 0.103 | 0.075 | 0.113 | -0.039 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| cdc25c |   | 1 | 164 | 153 | 168 | -0.025 | 0.187 | 0.084 | 0.267 | 0.070 | 0.059 | 0.367 | 0.125 | 0.204 | 0.250 | 0.052 | 0.054 | 0.351 |
|---|
| mapk14 |   | 1 | 164 | 153 | 168 | -0.062 | 0.058 | 0.135 | 0.204 | 0.086 | 0.061 | 0.389 | 0.068 | 0.246 | 0.068 | 0.056 | 0.011 | 0.192 |
|---|
| celsr3 |   | 1 | 164 | 153 | 168 | 0.097 | 0.124 | 0.198 | 0.213 | 0.100 | -0.091 | 0.450 | 0.148 | 0.106 | 0.103 | 0.030 | 0.069 | 0.126 |
|---|
| eif4g1 |   | 5 | 50 | 153 | 142 | 0.123 | 0.262 | 0.043 | 0.013 | 0.098 | 0.189 | 0.016 | 0.194 | 0.076 | 0.147 | -0.154 | -0.028 | 0.268 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| cks2 |   | 44 | 6 | 73 | 54 | 0.050 | 0.233 | 0.170 | 0.103 | 0.186 | 0.172 | 0.093 | 0.167 | 0.263 | 0.151 | 0.145 | 0.284 | 0.383 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| gcn1l1 |   | 1 | 164 | 153 | 168 | 0.022 | 0.081 | 0.125 | -0.072 | 0.113 | undef | undef | 0.092 | 0.011 | 0.208 | 0.044 | 0.030 | 0.333 |
|---|
| mars |   | 1 | 164 | 153 | 168 | 0.114 | 0.207 | 0.114 | 0.083 | 0.198 | 0.212 | -0.068 | 0.340 | 0.139 | 0.180 | 0.050 | 0.129 | 0.116 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
GO Enrichment output for subnetwork 10985-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.329E-08 | 3.056E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.787E-06 | 0.01010459 |
|---|
| interphase | GO:0051325 |  | 9.628E-06 | 7.382E-03 |
|---|
| regulation of translation | GO:0006417 |  | 1.599E-05 | 9.193E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 2.291E-05 | 0.01053646 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.891E-05 | 0.0149147 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.465E-05 | 0.01466918 |
|---|
| response to UV | GO:0009411 |  | 4.771E-05 | 0.01371691 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.801E-05 | 0.01226912 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.395E-05 | 0.01470891 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.395E-05 | 0.01337173 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.466E-10 | 2.068E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 1.291E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 5.29E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 8.959E-06 | 5.472E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.057E-05 | 5.164E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 4.55E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 5.831E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.682E-05 | 8.191E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.682E-05 | 7.281E-03 |
|---|
| thymus development | GO:0048538 |  | 3.277E-05 | 8.005E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.93E-05 | 8.729E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.587E-09 | 3.817E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 1.855E-03 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 1.401E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 7.727E-06 | 4.648E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 4.574E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.461E-05 | 5.858E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 5.021E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 6.496E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.468E-05 | 9.27E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.468E-05 | 8.343E-03 |
|---|
| thymus development | GO:0048538 |  | 4.236E-05 | 9.265E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.466E-10 | 2.068E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.057E-06 | 1.291E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 6.496E-06 | 5.29E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 8.959E-06 | 5.472E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.057E-05 | 5.164E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 4.55E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 5.831E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 2.682E-05 | 8.191E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 2.682E-05 | 7.281E-03 |
|---|
| thymus development | GO:0048538 |  | 3.277E-05 | 8.005E-03 |
|---|
| regulation of B cell proliferation | GO:0030888 |  | 3.93E-05 | 8.729E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.587E-09 | 3.817E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 1.855E-03 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 1.401E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 7.727E-06 | 4.648E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 4.574E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.461E-05 | 5.858E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 5.021E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 6.496E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.468E-05 | 9.27E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.468E-05 | 8.343E-03 |
|---|
| thymus development | GO:0048538 |  | 4.236E-05 | 9.265E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.198E-10 | 4.278E-07 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.62E-10 | 8.25E-07 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.997E-08 | 3.568E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 2.247E-07 | 2.006E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 4.756E-07 | 3.397E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 6.132E-07 | 3.65E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.61E-06 | 2.862E-03 |
|---|
| SAP kinase activity | GO:0016909 |  | 7.176E-06 | 3.203E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.969E-06 | 3.162E-03 |
|---|
| response to UV | GO:0009411 |  | 9.657E-06 | 3.449E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.076E-05 | 3.492E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 4.632E-10 | 1.69E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.407E-10 | 1.351E-06 |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 3.474E-08 | 4.224E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 3.24E-07 | 2.955E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 5.735E-07 | 4.184E-04 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.416E-07 | 4.509E-04 |
|---|
| interphase | GO:0051325 |  | 8.182E-07 | 4.264E-04 |
|---|
| regulation of translational initiation | GO:0006446 |  | 5.145E-06 | 2.346E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 7.506E-06 | 3.042E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 7.75E-06 | 2.827E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 7.75E-06 | 2.57E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.587E-09 | 3.817E-06 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.542E-06 | 1.855E-03 |
|---|
| interphase | GO:0051325 |  | 1.746E-06 | 1.401E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 7.727E-06 | 4.648E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.506E-06 | 4.574E-03 |
|---|
| regulation of translational initiation | GO:0006446 |  | 1.461E-05 | 5.858E-03 |
|---|
| response to UV | GO:0009411 |  | 1.461E-05 | 5.021E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.16E-05 | 6.496E-03 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 3.468E-05 | 9.27E-03 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 3.468E-05 | 8.343E-03 |
|---|
| thymus development | GO:0048538 |  | 4.236E-05 | 9.265E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.329E-08 | 3.056E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.787E-06 | 0.01010459 |
|---|
| interphase | GO:0051325 |  | 9.628E-06 | 7.382E-03 |
|---|
| regulation of translation | GO:0006417 |  | 1.599E-05 | 9.193E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 2.291E-05 | 0.01053646 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.891E-05 | 0.0149147 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.465E-05 | 0.01466918 |
|---|
| response to UV | GO:0009411 |  | 4.771E-05 | 0.01371691 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.801E-05 | 0.01226912 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.395E-05 | 0.01470891 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.395E-05 | 0.01337173 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.329E-08 | 3.056E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.787E-06 | 0.01010459 |
|---|
| interphase | GO:0051325 |  | 9.628E-06 | 7.382E-03 |
|---|
| regulation of translation | GO:0006417 |  | 1.599E-05 | 9.193E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 2.291E-05 | 0.01053646 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.891E-05 | 0.0149147 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.465E-05 | 0.01466918 |
|---|
| response to UV | GO:0009411 |  | 4.771E-05 | 0.01371691 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.801E-05 | 0.01226912 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.395E-05 | 0.01470891 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.395E-05 | 0.01337173 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.329E-08 | 3.056E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.787E-06 | 0.01010459 |
|---|
| interphase | GO:0051325 |  | 9.628E-06 | 7.382E-03 |
|---|
| regulation of translation | GO:0006417 |  | 1.599E-05 | 9.193E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 2.291E-05 | 0.01053646 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.891E-05 | 0.0149147 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.465E-05 | 0.01466918 |
|---|
| response to UV | GO:0009411 |  | 4.771E-05 | 0.01371691 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.801E-05 | 0.01226912 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.395E-05 | 0.01470891 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.395E-05 | 0.01337173 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.329E-08 | 3.056E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.787E-06 | 0.01010459 |
|---|
| interphase | GO:0051325 |  | 9.628E-06 | 7.382E-03 |
|---|
| regulation of translation | GO:0006417 |  | 1.599E-05 | 9.193E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 2.291E-05 | 0.01053646 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.891E-05 | 0.0149147 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.465E-05 | 0.01466918 |
|---|
| response to UV | GO:0009411 |  | 4.771E-05 | 0.01371691 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.801E-05 | 0.01226912 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 6.395E-05 | 0.01470891 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 6.395E-05 | 0.01337173 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.253E-08 | 2.973E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 7.463E-06 | 8.854E-03 |
|---|
| interphase | GO:0051325 |  | 8.169E-06 | 6.462E-03 |
|---|
| traversing start control point of mitotic cell cycle | GO:0007089 |  | 2.047E-05 | 0.01214657 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 3.546E-05 | 0.01683053 |
|---|
| regulation of translational initiation | GO:0006446 |  | 4.057E-05 | 0.0160442 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.292E-05 | 0.01454951 |
|---|
| response to UV | GO:0009411 |  | 4.329E-05 | 0.012841 |
|---|
| negative regulation of cyclin-dependent protein kinase activity | GO:0045736 |  | 5.717E-05 | 0.01507473 |
|---|
| positive regulation of B cell proliferation | GO:0030890 |  | 9.172E-05 | 0.02176509 |
|---|
| thymus development | GO:0048538 |  | 1.12E-04 | 0.02416156 |
|---|



