Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10736-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1037 | 7.338e-03 | 8.752e-03 | 7.841e-02 |
|---|
| IPC-NIBC-129 | 0.2541 | 5.663e-02 | 7.264e-02 | 4.903e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2633 | 1.824e-03 | 3.333e-03 | 8.488e-02 |
|---|
| Loi_GPL570 | 0.3220 | 5.812e-02 | 5.770e-02 | 2.844e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1450 | 7.000e-06 | 5.000e-05 | 4.900e-05 |
|---|
| Parker_GPL1390 | 0.2492 | 3.563e-01 | 3.854e-01 | 8.791e-01 |
|---|
| Parker_GPL887 | 0.0812 | 7.190e-01 | 7.434e-01 | 7.747e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3301 | 1.130e-01 | 8.428e-02 | 5.573e-01 |
|---|
| Schmidt | 0.2432 | 1.647e-02 | 2.301e-02 | 1.903e-01 |
|---|
| Sotiriou | 0.2847 | 2.087e-03 | 5.760e-04 | 5.466e-02 |
|---|
| Wang | 0.2180 | 1.406e-01 | 1.024e-01 | 4.536e-01 |
|---|
| Zhang | 0.2592 | 6.045e-02 | 3.585e-02 | 1.520e-01 |
|---|
| Zhou | 0.4237 | 8.139e-02 | 8.833e-02 | 4.756e-01 |
|---|
Expression data for subnetwork 10736-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
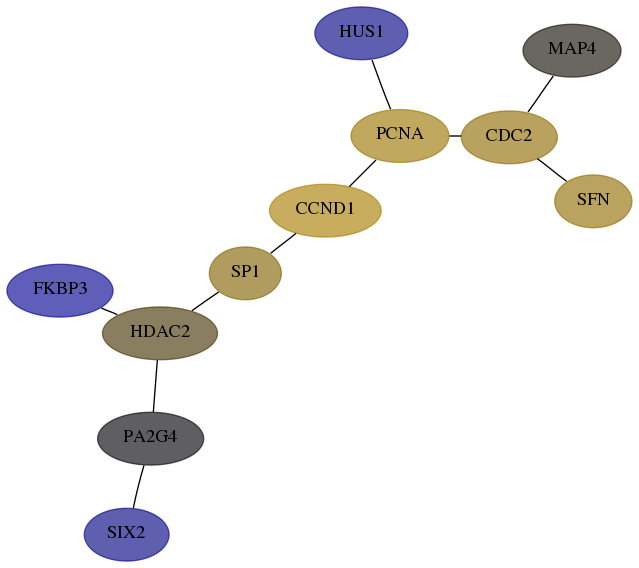
Score for each gene in subnetwork 10736-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| pcna |   | 6 | 43 | 203 | 180 | 0.096 | 0.202 | 0.109 | 0.180 | 0.118 | 0.119 | 0.104 | 0.256 | 0.151 | 0.059 | 0.162 | 0.123 | 0.233 |
|---|
| hus1 |   | 2 | 104 | 203 | 190 | -0.058 | 0.197 | 0.116 | 0.109 | 0.101 | 0.027 | 0.397 | 0.160 | 0.072 | 0.082 | 0.081 | 0.071 | 0.120 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| fkbp3 |   | 2 | 104 | 279 | 265 | -0.072 | 0.129 | 0.144 | 0.166 | 0.136 | -0.034 | -0.215 | 0.010 | 0.159 | 0.052 | 0.058 | 0.199 | 0.288 |
|---|
| sp1 |   | 4 | 60 | 203 | 183 | 0.054 | 0.186 | 0.147 | 0.242 | 0.125 | 0.033 | -0.370 | 0.202 | 0.284 | 0.227 | 0.194 | 0.091 | -0.068 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| map4 |   | 16 | 13 | 73 | 55 | 0.002 | 0.046 | 0.067 | -0.043 | 0.034 | 0.119 | 0.159 | -0.058 | -0.020 | 0.043 | 0.108 | -0.074 | 0.302 |
|---|
| hdac2 |   | 2 | 104 | 279 | 265 | 0.012 | 0.245 | 0.030 | 0.077 | 0.070 | 0.307 | -0.204 | 0.183 | 0.222 | -0.058 | 0.086 | 0.133 | 0.181 |
|---|
| pa2g4 |   | 1 | 164 | 279 | 286 | -0.001 | 0.092 | 0.146 | 0.101 | 0.134 | 0.030 | -0.023 | 0.212 | 0.129 | 0.108 | 0.032 | -0.012 | 0.179 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| six2 |   | 4 | 60 | 128 | 114 | -0.053 | 0.195 | 0.158 | 0.113 | 0.095 | 0.111 | 0.515 | -0.037 | 0.071 | 0.094 | 0.090 | -0.065 | -0.079 |
|---|
GO Enrichment output for subnetwork 10736-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.106E-05 | 0.07143888 |
|---|
| response to UV | GO:0009411 |  | 3.32E-05 | 0.03817708 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.777E-05 | 0.02171932 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.534E-05 | 0.02085546 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 5.032E-05 | 0.01928987 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 5.032E-05 | 0.01653418 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 6.008E-05 | 0.01727237 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 9.86E-05 | 0.02519704 |
|---|
| histone deacetylation | GO:0016575 |  | 9.86E-05 | 0.02267734 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.182E-04 | 0.02471817 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.975E-06 | 0.01459787 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.394E-06 | 9.032E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.959E-06 | 7.295E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 6.824E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.18E-05 | 5.766E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.245E-05 | 5.069E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.605E-05 | 5.601E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 5.102E-03 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 2.147E-05 | 5.828E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.277E-05 | 8.005E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 3.453E-05 | 7.67E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 8.321E-06 | 0.02001974 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.21E-05 | 0.0145595 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.247E-05 | 0.01000411 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 9.829E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.93E-05 | 9.287E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.036E-05 | 8.164E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 7.994E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.377E-05 | 7.148E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.989E-05 | 7.989E-03 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 2.989E-05 | 7.191E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 5.225E-05 | 0.01142822 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 5.975E-06 | 0.01459787 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 7.394E-06 | 9.032E-03 |
|---|
| establishment of spindle localization | GO:0051293 |  | 8.959E-06 | 7.295E-03 |
|---|
| response to UV | GO:0009411 |  | 1.117E-05 | 6.824E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.18E-05 | 5.766E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.245E-05 | 5.069E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 1.605E-05 | 5.601E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.671E-05 | 5.102E-03 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 2.147E-05 | 5.828E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 3.277E-05 | 8.005E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 3.453E-05 | 7.67E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 8.321E-06 | 0.02001974 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.21E-05 | 0.0145595 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.247E-05 | 0.01000411 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 9.829E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.93E-05 | 9.287E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.036E-05 | 8.164E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 7.994E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.377E-05 | 7.148E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.989E-05 | 7.989E-03 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 2.989E-05 | 7.191E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 5.225E-05 | 0.01142822 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.259E-08 | 4.494E-05 |
|---|
| replication fork | GO:0005657 |  | 1.002E-06 | 1.789E-03 |
|---|
| response to UV | GO:0009411 |  | 4.081E-06 | 4.857E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 4.142E-06 | 3.697E-03 |
|---|
| MutLalpha complex binding | GO:0032405 |  | 4.142E-06 | 2.958E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.339E-06 | 2.583E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.339E-06 | 2.214E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 4.889E-06 | 2.183E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 5.181E-06 | 2.056E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 6.21E-06 | 2.218E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 6.21E-06 | 2.016E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.125E-08 | 4.104E-05 |
|---|
| replication fork | GO:0005657 |  | 7.285E-07 | 1.329E-03 |
|---|
| response to UV | GO:0009411 |  | 2.813E-06 | 3.42E-03 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 3.763E-06 | 3.432E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.763E-06 | 2.745E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 3.878E-06 | 2.358E-03 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 3.878E-06 | 2.021E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.119E-06 | 1.878E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 4.63E-06 | 1.877E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.642E-06 | 2.058E-03 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 5.642E-06 | 1.871E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| meiotic chromosome segregation | GO:0045132 |  | 8.321E-06 | 0.02001974 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.21E-05 | 0.0145595 |
|---|
| establishment of spindle localization | GO:0051293 |  | 1.247E-05 | 0.01000411 |
|---|
| response to UV | GO:0009411 |  | 1.634E-05 | 9.829E-03 |
|---|
| regulation of DNA replication | GO:0006275 |  | 1.93E-05 | 9.287E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 2.036E-05 | 8.164E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.326E-05 | 7.994E-03 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 2.377E-05 | 7.148E-03 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 2.989E-05 | 7.989E-03 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 2.989E-05 | 7.191E-03 |
|---|
| negative regulation of organelle organization | GO:0010639 |  | 5.225E-05 | 0.01142822 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.106E-05 | 0.07143888 |
|---|
| response to UV | GO:0009411 |  | 3.32E-05 | 0.03817708 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.777E-05 | 0.02171932 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.534E-05 | 0.02085546 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 5.032E-05 | 0.01928987 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 5.032E-05 | 0.01653418 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 6.008E-05 | 0.01727237 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 9.86E-05 | 0.02519704 |
|---|
| histone deacetylation | GO:0016575 |  | 9.86E-05 | 0.02267734 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.182E-04 | 0.02471817 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.106E-05 | 0.07143888 |
|---|
| response to UV | GO:0009411 |  | 3.32E-05 | 0.03817708 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.777E-05 | 0.02171932 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.534E-05 | 0.02085546 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 5.032E-05 | 0.01928987 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 5.032E-05 | 0.01653418 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 6.008E-05 | 0.01727237 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 9.86E-05 | 0.02519704 |
|---|
| histone deacetylation | GO:0016575 |  | 9.86E-05 | 0.02267734 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.182E-04 | 0.02471817 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.106E-05 | 0.07143888 |
|---|
| response to UV | GO:0009411 |  | 3.32E-05 | 0.03817708 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.777E-05 | 0.02171932 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.534E-05 | 0.02085546 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 5.032E-05 | 0.01928987 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 5.032E-05 | 0.01653418 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 6.008E-05 | 0.01727237 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 9.86E-05 | 0.02519704 |
|---|
| histone deacetylation | GO:0016575 |  | 9.86E-05 | 0.02267734 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.182E-04 | 0.02471817 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 3.106E-05 | 0.07143888 |
|---|
| response to UV | GO:0009411 |  | 3.32E-05 | 0.03817708 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.776E-05 | 0.02894696 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.777E-05 | 0.02171932 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.534E-05 | 0.02085546 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 5.032E-05 | 0.01928987 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 5.032E-05 | 0.01653418 |
|---|
| DNA damage response. signal transduction | GO:0042770 |  | 6.008E-05 | 0.01727237 |
|---|
| double-strand break repair via homologous recombination | GO:0000724 |  | 9.86E-05 | 0.02519704 |
|---|
| histone deacetylation | GO:0016575 |  | 9.86E-05 | 0.02267734 |
|---|
| positive regulation of cell growth | GO:0030307 |  | 1.182E-04 | 0.02471817 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| definitive hemopoiesis | GO:0060216 |  | 1.514E-05 | 0.03591565 |
|---|
| embryonic camera-type eye development | GO:0031076 |  | 2.269E-05 | 0.02691597 |
|---|
| embryonic skeletal system development | GO:0048706 |  | 2.743E-05 | 0.02169738 |
|---|
| response to UV | GO:0009411 |  | 2.743E-05 | 0.01627304 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.173E-05 | 0.01506132 |
|---|
| enucleate erythrocyte differentiation | GO:0043353 |  | 3.173E-05 | 0.0125511 |
|---|
| regulation of DNA replication | GO:0006275 |  | 3.723E-05 | 0.01262131 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 3.943E-05 | 0.01169656 |
|---|
| keratinocyte proliferation | GO:0043616 |  | 4.228E-05 | 0.01114794 |
|---|
| middle ear morphogenesis | GO:0042474 |  | 4.228E-05 | 0.01003314 |
|---|
| trophectodermal cell differentiation | GO:0001829 |  | 4.228E-05 | 9.121E-03 |
|---|



