Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10656-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2311 | 6.773e-03 | 8.072e-03 | 7.375e-02 |
|---|
| IPC-NIBC-129 | 0.2743 | 9.081e-02 | 1.192e-01 | 6.421e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2672 | 5.800e-04 | 1.190e-03 | 3.572e-02 |
|---|
| Loi_GPL570 | 0.3746 | 4.403e-02 | 4.369e-02 | 2.318e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1703 | 1.960e-04 | 8.940e-04 | 5.243e-03 |
|---|
| Parker_GPL1390 | 0.2145 | 6.351e-02 | 6.074e-02 | 4.294e-01 |
|---|
| Parker_GPL887 | 0.0951 | 6.670e-01 | 6.943e-01 | 7.205e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3211 | 3.263e-01 | 2.870e-01 | 8.867e-01 |
|---|
| Schmidt | 0.1957 | 9.239e-03 | 1.324e-02 | 1.230e-01 |
|---|
| Sotiriou | 0.3200 | 1.990e-03 | 5.420e-04 | 5.280e-02 |
|---|
| Wang | 0.1634 | 9.378e-02 | 6.380e-02 | 3.505e-01 |
|---|
| Zhang | 0.1517 | 1.328e-01 | 9.193e-02 | 3.074e-01 |
|---|
| Zhou | 0.4266 | 8.859e-02 | 9.685e-02 | 5.065e-01 |
|---|
Expression data for subnetwork 10656-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
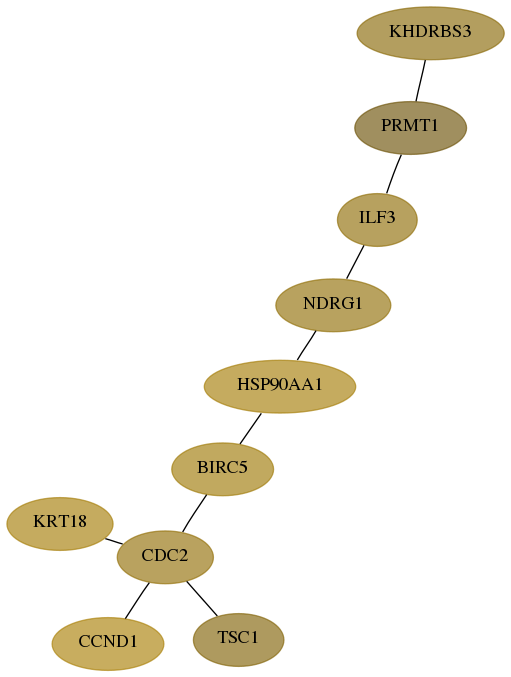
Score for each gene in subnetwork 10656-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| krt18 |   | 15 | 15 | 230 | 195 | 0.112 | -0.078 | 0.147 | 0.150 | 0.161 | -0.086 | -0.204 | 0.083 | -0.099 | 0.118 | 0.086 | -0.114 | 0.222 |
|---|
| ilf3 |   | 2 | 104 | 64 | 71 | 0.069 | 0.235 | 0.115 | 0.194 | 0.073 | 0.201 | 0.067 | 0.145 | 0.055 | 0.030 | -0.100 | 0.013 | 0.265 |
|---|
| hsp90aa1 |   | 11 | 21 | 234 | 204 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 | 0.399 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| ndrg1 |   | 9 | 27 | 250 | 223 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.102 | 0.240 | 0.083 | 0.068 | 0.064 | 0.128 | 0.092 | -0.255 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| prmt1 |   | 2 | 104 | 64 | 71 | 0.029 | 0.172 | 0.254 | 0.122 | 0.144 | 0.192 | -0.083 | 0.169 | 0.003 | 0.154 | -0.065 | -0.147 | 0.219 |
|---|
| khdrbs3 |   | 3 | 79 | 64 | 68 | 0.060 | 0.275 | -0.073 | 0.091 | 0.049 | 0.150 | 0.116 | 0.145 | 0.171 | 0.054 | -0.038 | 0.134 | 0.140 |
|---|
| birc5 |   | 14 | 17 | 162 | 140 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | 0.124 | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 10656-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 0.02854035 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 0.01427018 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 0.01350943 |
|---|
| protein refolding | GO:0042026 |  | 1.86E-05 | 0.01069529 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.86E-05 | 8.556E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.86E-05 | 7.13E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 8.55E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.602E-05 | 7.482E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.602E-05 | 6.65E-03 |
|---|
| mast cell activation | GO:0045576 |  | 2.602E-05 | 5.985E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.602E-05 | 5.441E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 9.773E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.016E-06 | 4.906E-03 |
|---|
| protein refolding | GO:0042026 |  | 5.998E-06 | 4.885E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.998E-06 | 3.664E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 8.394E-06 | 4.102E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 8.394E-06 | 3.418E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 8.394E-06 | 2.93E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 8.394E-06 | 2.563E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.119E-05 | 3.037E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.119E-05 | 2.733E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.119E-05 | 2.485E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 0.01223916 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 6.12E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.728E-06 | 4.594E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.627E-06 | 4.588E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.067E-05 | 5.136E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.067E-05 | 4.28E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.067E-05 | 3.668E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.067E-05 | 3.21E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.803E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.422E-05 | 3.422E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.422E-05 | 3.111E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 4.001E-06 | 9.773E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.016E-06 | 4.906E-03 |
|---|
| protein refolding | GO:0042026 |  | 5.998E-06 | 4.885E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.998E-06 | 3.664E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 8.394E-06 | 4.102E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 8.394E-06 | 3.418E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 8.394E-06 | 2.93E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 8.394E-06 | 2.563E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.119E-05 | 3.037E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.119E-05 | 2.733E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.119E-05 | 2.485E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 0.01223916 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 6.12E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.728E-06 | 4.594E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.627E-06 | 4.588E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.067E-05 | 5.136E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.067E-05 | 4.28E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.067E-05 | 3.668E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.067E-05 | 3.21E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.803E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.422E-05 | 3.422E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.422E-05 | 3.111E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.578E-06 | 9.205E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 2.578E-06 | 4.602E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 2.578E-06 | 3.068E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 3.865E-06 | 3.451E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 3.865E-06 | 2.761E-03 |
|---|
| mast cell activation | GO:0045576 |  | 3.865E-06 | 2.301E-03 |
|---|
| response to metal ion | GO:0010038 |  | 4.517E-06 | 2.305E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 4.731E-06 | 2.112E-03 |
|---|
| regulation of phosphoprotein phosphatase activity | GO:0043666 |  | 5.41E-06 | 2.147E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 5.41E-06 | 1.932E-03 |
|---|
| centriolar satellite | GO:0034451 |  | 5.41E-06 | 1.756E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| midbody | GO:0030496 |  | 3.311E-08 | 1.208E-04 |
|---|
| spindle microtubule | GO:0005876 |  | 1.226E-07 | 2.236E-04 |
|---|
| chaperone binding | GO:0051087 |  | 1.711E-07 | 2.081E-04 |
|---|
| microtubule organizing center part | GO:0044450 |  | 7.417E-07 | 6.764E-04 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.839E-06 | 1.342E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 3.136E-06 | 1.907E-03 |
|---|
| nitric-oxide synthase regulator activity | GO:0030235 |  | 3.136E-06 | 1.634E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 3.136E-06 | 1.43E-03 |
|---|
| positive regulation of cell-matrix adhesion | GO:0001954 |  | 3.136E-06 | 1.271E-03 |
|---|
| RNA polymerase II carboxy-terminal domain kinase activity | GO:0008353 |  | 3.136E-06 | 1.144E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 4.703E-06 | 1.56E-03 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 5.087E-06 | 0.01223916 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 5.087E-06 | 6.12E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 5.728E-06 | 4.594E-03 |
|---|
| protein refolding | GO:0042026 |  | 7.627E-06 | 4.588E-03 |
|---|
| tumor necrosis factor-mediated signaling pathway | GO:0033209 |  | 1.067E-05 | 5.136E-03 |
|---|
| Golgi to plasma membrane transport | GO:0006893 |  | 1.067E-05 | 4.28E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.067E-05 | 3.668E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.067E-05 | 3.21E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 1.422E-05 | 3.803E-03 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.422E-05 | 3.422E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.422E-05 | 3.111E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 0.02854035 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 0.01427018 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 0.01350943 |
|---|
| protein refolding | GO:0042026 |  | 1.86E-05 | 0.01069529 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.86E-05 | 8.556E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.86E-05 | 7.13E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 8.55E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.602E-05 | 7.482E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.602E-05 | 6.65E-03 |
|---|
| mast cell activation | GO:0045576 |  | 2.602E-05 | 5.985E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.602E-05 | 5.441E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 0.02854035 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 0.01427018 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 0.01350943 |
|---|
| protein refolding | GO:0042026 |  | 1.86E-05 | 0.01069529 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.86E-05 | 8.556E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.86E-05 | 7.13E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 8.55E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.602E-05 | 7.482E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.602E-05 | 6.65E-03 |
|---|
| mast cell activation | GO:0045576 |  | 2.602E-05 | 5.985E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.602E-05 | 5.441E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 0.02854035 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 0.01427018 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 0.01350943 |
|---|
| protein refolding | GO:0042026 |  | 1.86E-05 | 0.01069529 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.86E-05 | 8.556E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.86E-05 | 7.13E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 8.55E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.602E-05 | 7.482E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.602E-05 | 6.65E-03 |
|---|
| mast cell activation | GO:0045576 |  | 2.602E-05 | 5.985E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.602E-05 | 5.441E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.241E-05 | 0.02854035 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 1.241E-05 | 0.01427018 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.762E-05 | 0.01350943 |
|---|
| protein refolding | GO:0042026 |  | 1.86E-05 | 0.01069529 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.86E-05 | 8.556E-03 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 1.86E-05 | 7.13E-03 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 2.602E-05 | 8.55E-03 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 2.602E-05 | 7.482E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.602E-05 | 6.65E-03 |
|---|
| mast cell activation | GO:0045576 |  | 2.602E-05 | 5.985E-03 |
|---|
| regulation of focal adhesion formation | GO:0051893 |  | 2.602E-05 | 5.441E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein refolding | GO:0042026 |  | 2.345E-08 | 5.564E-05 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 4.1E-08 | 4.865E-05 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 6.555E-08 | 5.185E-05 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 1.323E-06 | 7.849E-04 |
|---|
| response to unfolded protein | GO:0006986 |  | 1.68E-06 | 7.974E-04 |
|---|
| response to protein stimulus | GO:0051789 |  | 2.143E-06 | 8.477E-04 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 2.342E-06 | 7.94E-04 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 2.66E-06 | 7.89E-04 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.166E-05 | 3.074E-03 |
|---|
| short-chain fatty acid metabolic process | GO:0046459 |  | 1.166E-05 | 2.766E-03 |
|---|
| galactolipid metabolic process | GO:0019374 |  | 1.166E-05 | 2.515E-03 |
|---|



