Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10607-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2445 | 8.296e-03 | 9.904e-03 | 8.608e-02 |
|---|
| IPC-NIBC-129 | 0.2573 | 4.858e-02 | 6.173e-02 | 4.438e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2573 | 6.420e-04 | 1.305e-03 | 3.870e-02 |
|---|
| Loi_GPL570 | 0.3699 | 5.567e-02 | 5.527e-02 | 2.757e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1473 | 1.000e-06 | 6.000e-06 | 1.000e-06 |
|---|
| Parker_GPL1390 | 0.2604 | 5.249e-02 | 4.927e-02 | 3.872e-01 |
|---|
| Parker_GPL887 | 0.2815 | 2.096e-01 | 2.399e-01 | 2.008e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3861 | 1.957e-01 | 1.592e-01 | 7.377e-01 |
|---|
| Schmidt | 0.2758 | 9.744e-03 | 1.394e-02 | 1.282e-01 |
|---|
| Sotiriou | 0.3167 | 1.907e-03 | 5.140e-04 | 5.118e-02 |
|---|
| Wang | 0.1449 | 1.356e-01 | 9.817e-02 | 4.437e-01 |
|---|
| Zhang | 0.2046 | 1.723e-01 | 1.256e-01 | 3.820e-01 |
|---|
| Zhou | 0.4291 | 4.775e-02 | 4.928e-02 | 3.041e-01 |
|---|
Expression data for subnetwork 10607-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
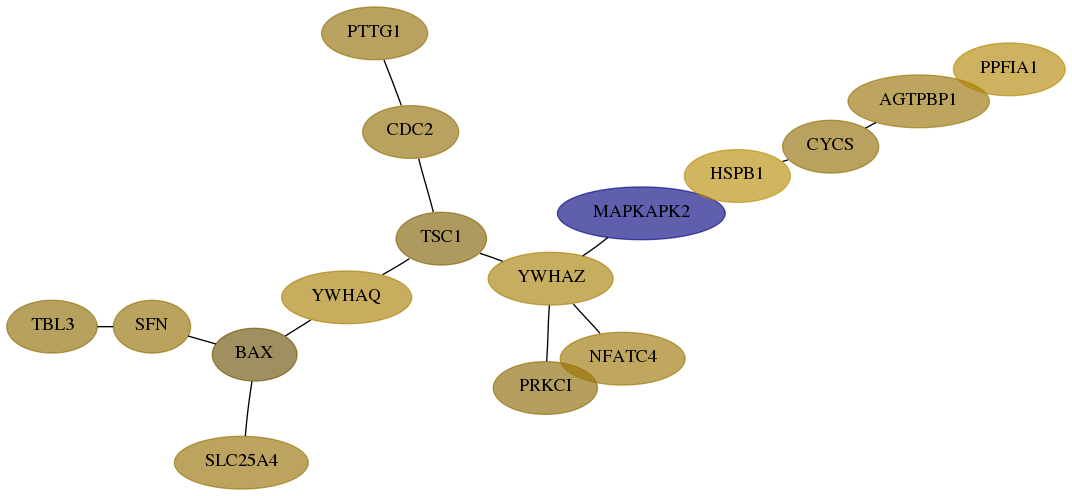
Score for each gene in subnetwork 10607-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| agtpbp1 |   | 11 | 21 | 17 | 13 | 0.086 | 0.219 | 0.009 | 0.181 | 0.071 | 0.078 | 0.296 | 0.026 | 0.110 | 0.151 | 0.103 | 0.064 | 0.226 |
|---|
| tbl3 |   | 4 | 60 | 31 | 30 | 0.068 | -0.007 | 0.137 | 0.048 | 0.104 | -0.071 | 0.518 | 0.154 | 0.067 | 0.128 | 0.061 | 0.054 | 0.267 |
|---|
| nfatc4 |   | 8 | 33 | 31 | 25 | 0.094 | -0.056 | -0.021 | 0.204 | 0.079 | -0.008 | 0.243 | 0.060 | 0.077 | -0.012 | 0.080 | 0.131 | 0.154 |
|---|
| cycs |   | 10 | 25 | 17 | 14 | 0.073 | 0.206 | 0.080 | 0.094 | 0.117 | -0.056 | 0.202 | 0.272 | 0.157 | 0.077 | 0.190 | 0.135 | 0.234 |
|---|
| slc25a4 |   | 3 | 79 | 17 | 22 | 0.084 | 0.041 | 0.064 | 0.086 | 0.187 | 0.117 | 0.044 | 0.166 | 0.004 | 0.178 | 0.108 | 0.025 | 0.188 |
|---|
| pttg1 |   | 10 | 25 | 31 | 21 | 0.072 | 0.270 | 0.175 | 0.183 | 0.200 | 0.208 | 0.347 | 0.334 | 0.224 | 0.204 | 0.147 | 0.210 | 0.309 |
|---|
| ywhaz |   | 11 | 21 | 31 | 20 | 0.124 | 0.089 | 0.087 | 0.200 | 0.141 | 0.110 | 0.228 | 0.199 | 0.056 | 0.123 | -0.026 | 0.103 | 0.213 |
|---|
| bax |   | 9 | 27 | 17 | 15 | 0.029 | 0.185 | 0.233 | 0.192 | 0.088 | -0.043 | 0.184 | 0.172 | 0.175 | 0.019 | -0.094 | 0.041 | 0.235 |
|---|
| ppfia1 |   | 8 | 33 | 17 | 16 | 0.164 | 0.080 | 0.149 | 0.269 | 0.151 | 0.214 | 0.093 | 0.225 | 0.036 | 0.176 | 0.153 | 0.127 | 0.272 |
|---|
| prkci |   | 12 | 20 | 31 | 19 | 0.065 | 0.104 | 0.012 | 0.170 | 0.112 | 0.088 | -0.103 | -0.170 | 0.196 | 0.128 | -0.054 | 0.124 | 0.149 |
|---|
| hspb1 |   | 19 | 11 | 17 | 11 | 0.190 | -0.050 | 0.184 | 0.212 | 0.231 | -0.026 | -0.065 | 0.289 | -0.021 | 0.201 | 0.125 | -0.077 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| ywhaq |   | 3 | 79 | 31 | 38 | 0.129 | 0.110 | 0.049 | 0.116 | 0.071 | 0.160 | -0.210 | 0.038 | 0.007 | 0.054 | -0.032 | 0.138 | 0.187 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| mapkapk2 |   | 13 | 19 | 17 | 12 | -0.050 | 0.102 | 0.156 | -0.116 | 0.090 | 0.168 | 0.303 | 0.074 | 0.171 | 0.117 | 0.085 | -0.124 | 0.312 |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
GO Enrichment output for subnetwork 10607-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.037E-08 | 1.158E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.251E-07 | 9.488E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.783E-06 | 2.134E-03 |
|---|
| regulation of caspase activity | GO:0043281 |  | 5.078E-06 | 2.92E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.639E-06 | 2.594E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 5.734E-06 | 2.198E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 7.65E-06 | 2.513E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.964E-06 | 2.865E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.788E-05 | 4.569E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.418E-05 | 5.562E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.911E-05 | 6.086E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 9.528E-09 | 2.328E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.222E-08 | 1.493E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.496E-08 | 1.218E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.058E-08 | 1.257E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 5.324E-08 | 2.601E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.974E-07 | 8.036E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.71E-07 | 9.457E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.368E-07 | 1.029E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.295E-07 | 1.437E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.425E-07 | 1.57E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.253E-06 | 2.783E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.566E-08 | 3.767E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.413E-08 | 2.902E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.773E-08 | 2.224E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.863E-08 | 2.324E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.747E-08 | 4.209E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.221E-07 | 1.292E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.449E-07 | 1.529E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.613E-07 | 1.688E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.69E-07 | 2.323E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.054E-06 | 2.537E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.055E-06 | 4.496E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 9.528E-09 | 2.328E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 1.222E-08 | 1.493E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 1.496E-08 | 1.218E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 2.058E-08 | 1.257E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 5.324E-08 | 2.601E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 1.974E-07 | 8.036E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 2.71E-07 | 9.457E-05 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 3.368E-07 | 1.029E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 5.295E-07 | 1.437E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 6.425E-07 | 1.57E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 1.253E-06 | 2.783E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.566E-08 | 3.767E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.413E-08 | 2.902E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.773E-08 | 2.224E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.863E-08 | 2.324E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.747E-08 | 4.209E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.221E-07 | 1.292E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.449E-07 | 1.529E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.613E-07 | 1.688E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.69E-07 | 2.323E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.054E-06 | 2.537E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.055E-06 | 4.496E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| regulation of caspase activity | GO:0043281 |  | 6.821E-07 | 2.436E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 7.683E-07 | 1.372E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 1.019E-06 | 1.213E-03 |
|---|
| regulation of Ras GTPase activity | GO:0032318 |  | 1.395E-06 | 1.246E-03 |
|---|
| regulation of GTPase activity | GO:0043087 |  | 3.023E-06 | 2.159E-03 |
|---|
| cysteine-type endopeptidase inhibitor activity | GO:0004869 |  | 4.19E-06 | 2.494E-03 |
|---|
| regulation of Rab protein signal transduction | GO:0032483 |  | 1.013E-05 | 5.167E-03 |
|---|
| Rab GTPase activator activity | GO:0005097 |  | 1.092E-05 | 4.875E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.104E-05 | 4.378E-03 |
|---|
| mitochondrial genome maintenance | GO:0000002 |  | 1.104E-05 | 3.941E-03 |
|---|
| small-subunit processome | GO:0032040 |  | 1.104E-05 | 3.582E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 3.094E-08 | 1.129E-04 |
|---|
| release of cytochrome c from mitochondria | GO:0001836 |  | 3.204E-07 | 5.844E-04 |
|---|
| regulation of caspase activity | GO:0043281 |  | 3.565E-07 | 4.334E-04 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.346E-07 | 3.963E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 4.923E-07 | 3.592E-04 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 5.578E-07 | 3.391E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 9.995E-07 | 5.209E-04 |
|---|
| apoptotic mitochondrial changes | GO:0008637 |  | 1.165E-06 | 5.314E-04 |
|---|
| apoptotic nuclear changes | GO:0030262 |  | 1.165E-06 | 4.724E-04 |
|---|
| mitochondrion organization | GO:0007005 |  | 1.261E-06 | 4.599E-04 |
|---|
| DNA catabolic process. endonucleolytic | GO:0000737 |  | 1.349E-06 | 4.472E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 1.566E-08 | 3.767E-05 |
|---|
| regulation of caspase activity | GO:0043281 |  | 2.413E-08 | 2.902E-05 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 2.773E-08 | 2.224E-05 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 3.863E-08 | 2.324E-05 |
|---|
| activation of caspase activity by cytochrome c | GO:0008635 |  | 8.747E-08 | 4.209E-05 |
|---|
| activation of caspase activity | GO:0006919 |  | 3.221E-07 | 1.292E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 4.449E-07 | 1.529E-04 |
|---|
| positive regulation of caspase activity | GO:0043280 |  | 5.613E-07 | 1.688E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 8.69E-07 | 2.323E-04 |
|---|
| DNA fragmentation involved in apoptosis | GO:0006309 |  | 1.054E-06 | 2.537E-04 |
|---|
| cell structure disassembly during apoptosis | GO:0006921 |  | 2.055E-06 | 4.496E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.037E-08 | 1.158E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.251E-07 | 9.488E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.783E-06 | 2.134E-03 |
|---|
| regulation of caspase activity | GO:0043281 |  | 5.078E-06 | 2.92E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.639E-06 | 2.594E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 5.734E-06 | 2.198E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 7.65E-06 | 2.513E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.964E-06 | 2.865E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.788E-05 | 4.569E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.418E-05 | 5.562E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.911E-05 | 6.086E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.037E-08 | 1.158E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.251E-07 | 9.488E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.783E-06 | 2.134E-03 |
|---|
| regulation of caspase activity | GO:0043281 |  | 5.078E-06 | 2.92E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.639E-06 | 2.594E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 5.734E-06 | 2.198E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 7.65E-06 | 2.513E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.964E-06 | 2.865E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.788E-05 | 4.569E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.418E-05 | 5.562E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.911E-05 | 6.086E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.037E-08 | 1.158E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.251E-07 | 9.488E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.783E-06 | 2.134E-03 |
|---|
| regulation of caspase activity | GO:0043281 |  | 5.078E-06 | 2.92E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.639E-06 | 2.594E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 5.734E-06 | 2.198E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 7.65E-06 | 2.513E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.964E-06 | 2.865E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.788E-05 | 4.569E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.418E-05 | 5.562E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.911E-05 | 6.086E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 5.037E-08 | 1.158E-04 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.251E-07 | 9.488E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.783E-06 | 2.134E-03 |
|---|
| regulation of caspase activity | GO:0043281 |  | 5.078E-06 | 2.92E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.639E-06 | 2.594E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 5.734E-06 | 2.198E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 7.65E-06 | 2.513E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.964E-06 | 2.865E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.788E-05 | 4.569E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.418E-05 | 5.562E-03 |
|---|
| polyol metabolic process | GO:0019751 |  | 2.911E-05 | 6.086E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| polyol catabolic process | GO:0046174 |  | 3.776E-08 | 8.959E-05 |
|---|
| cellular carbohydrate catabolic process | GO:0044275 |  | 8.243E-07 | 9.78E-04 |
|---|
| fructose metabolic process | GO:0006000 |  | 2.534E-06 | 2.004E-03 |
|---|
| regulation of caspase activity | GO:0043281 |  | 4.181E-06 | 2.481E-03 |
|---|
| regulation of endopeptidase activity | GO:0052548 |  | 4.697E-06 | 2.229E-03 |
|---|
| alditol metabolic process | GO:0019400 |  | 5.707E-06 | 2.257E-03 |
|---|
| regulation of peptidase activity | GO:0052547 |  | 6.191E-06 | 2.099E-03 |
|---|
| sperm motility | GO:0030317 |  | 9.593E-06 | 2.846E-03 |
|---|
| hexose biosynthetic process | GO:0019319 |  | 1.65E-05 | 4.349E-03 |
|---|
| monosaccharide biosynthetic process | GO:0046364 |  | 2.189E-05 | 5.194E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.501E-05 | 5.394E-03 |
|---|



