Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10101-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.2091 | 9.668e-03 | 1.155e-02 | 9.661e-02 |
|---|
| IPC-NIBC-129 | 0.2315 | 8.617e-02 | 1.129e-01 | 6.250e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2499 | 7.925e-03 | 1.249e-02 | 2.286e-01 |
|---|
| Loi_GPL570 | 0.2538 | 7.903e-02 | 7.852e-02 | 3.532e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1619 | 3.000e-06 | 2.600e-05 | 1.500e-05 |
|---|
| Parker_GPL1390 | 0.2183 | 8.656e-02 | 8.519e-02 | 5.039e-01 |
|---|
| Parker_GPL887 | 0.1290 | 5.498e-01 | 5.821e-01 | 5.919e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3722 | 1.573e-01 | 1.236e-01 | 6.659e-01 |
|---|
| Schmidt | 0.2539 | 1.485e-02 | 2.085e-02 | 1.764e-01 |
|---|
| Sotiriou | 0.2910 | 1.621e-02 | 7.604e-03 | 2.210e-01 |
|---|
| Wang | 0.1974 | 1.075e-01 | 7.480e-02 | 3.829e-01 |
|---|
| Zhang | 0.2266 | 8.156e-02 | 5.131e-02 | 2.002e-01 |
|---|
| Zhou | 0.2991 | 5.457e-02 | 5.707e-02 | 3.428e-01 |
|---|
Expression data for subnetwork 10101-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
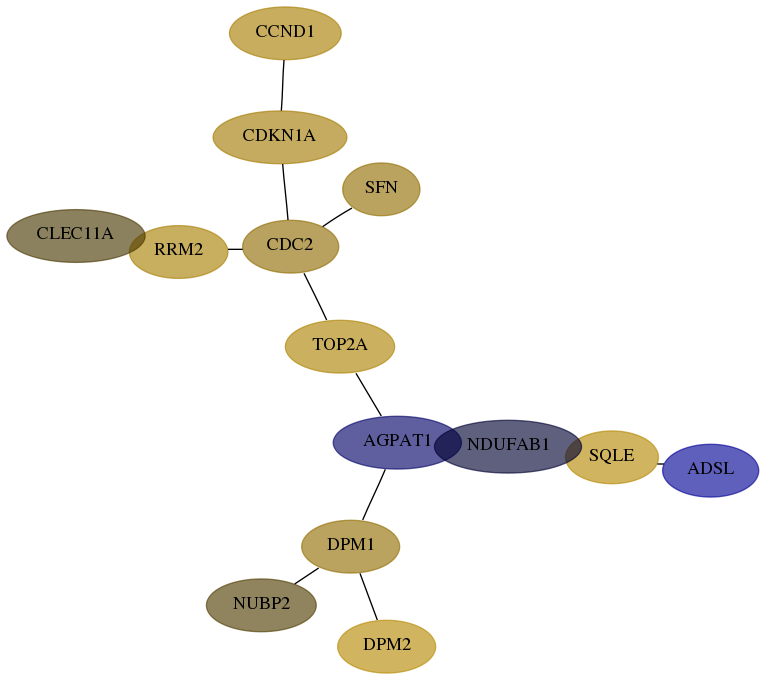
Score for each gene in subnetwork 10101-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| sqle |   | 1 | 164 | 363 | 363 | 0.174 | 0.149 | 0.034 | 0.168 | 0.145 | 0.049 | 0.203 | 0.326 | 0.125 | 0.077 | 0.077 | 0.173 | 0.171 |
|---|
| dpm2 |   | 1 | 164 | 363 | 363 | 0.176 | 0.032 | 0.117 | 0.096 | 0.111 | undef | 0.065 | 0.203 | 0.102 | 0.163 | -0.015 | 0.104 | 0.125 |
|---|
| rrm2 |   | 21 | 10 | 44 | 26 | 0.131 | 0.207 | 0.227 | 0.187 | 0.161 | 0.178 | 0.137 | 0.303 | 0.190 | 0.219 | 0.117 | 0.129 | 0.302 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| dpm1 |   | 1 | 164 | 363 | 363 | 0.078 | 0.094 | 0.010 | 0.065 | 0.078 | 0.115 | -0.055 | 0.094 | 0.196 | 0.023 | 0.102 | -0.025 | 0.078 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| ndufab1 |   | 2 | 104 | 319 | 306 | -0.007 | 0.205 | 0.083 | 0.106 | 0.157 | 0.038 | -0.073 | 0.175 | 0.193 | 0.142 | 0.112 | 0.031 | 0.138 |
|---|
| nubp2 |   | 1 | 164 | 363 | 363 | 0.016 | 0.076 | 0.089 | 0.208 | 0.090 | 0.015 | 0.325 | 0.090 | 0.081 | -0.006 | 0.074 | 0.197 | 0.080 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| agpat1 |   | 3 | 79 | 192 | 178 | -0.027 | 0.104 | 0.123 | 0.096 | 0.093 | 0.167 | 0.097 | 0.028 | 0.097 | 0.101 | -0.079 | 0.018 | -0.007 |
|---|
| adsl |   | 1 | 164 | 363 | 363 | -0.083 | 0.166 | 0.156 | 0.145 | 0.082 | 0.120 | 0.195 | 0.168 | 0.212 | 0.028 | 0.030 | 0.178 | undef |
|---|
| sfn |   | 50 | 4 | 29 | 17 | 0.077 | 0.137 | 0.111 | 0.172 | 0.112 | 0.119 | 0.141 | 0.219 | 0.100 | 0.147 | 0.025 | 0.079 | 0.181 |
|---|
| top2a |   | 11 | 21 | 93 | 76 | 0.147 | 0.183 | 0.185 | 0.100 | 0.206 | 0.014 | 0.035 | 0.309 | 0.261 | 0.183 | 0.072 | 0.234 | 0.273 |
|---|
| clec11a |   | 17 | 12 | 44 | 27 | 0.013 | 0.101 | 0.071 | -0.085 | 0.066 | 0.082 | 0.209 | -0.126 | -0.034 | -0.001 | 0.191 | 0.059 | -0.075 |
|---|
GO Enrichment output for subnetwork 10101-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.071E-08 | 2.463E-05 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 2.025E-08 | 2.328E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.152E-08 | 3.95E-05 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 5.892E-08 | 3.388E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 9.008E-08 | 4.143E-05 |
|---|
| glycerophospholipid metabolic process | GO:0006650 |  | 1.768E-07 | 6.778E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 2.552E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.076E-06 | 5.968E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 2.898E-06 | 7.406E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.378E-06 | 7.769E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 6.572E-06 | 1.374E-03 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.031E-09 | 2.518E-06 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 2.022E-09 | 2.469E-06 |
|---|
| phosphatidic acid metabolic process | GO:0046473 |  | 3.471E-09 | 2.826E-06 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 7.714E-09 | 4.711E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.651E-08 | 4.227E-05 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 6.961E-07 | 2.834E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 1.254E-06 | 4.375E-04 |
|---|
| GPI anchor biosynthetic process | GO:0006506 |  | 2.048E-06 | 6.254E-04 |
|---|
| GPI anchor metabolic process | GO:0006505 |  | 2.239E-06 | 6.077E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.655E-06 | 6.487E-04 |
|---|
| phosphoinositide biosynthetic process | GO:0046489 |  | 3.636E-06 | 8.075E-04 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.835E-09 | 4.415E-06 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 3.451E-09 | 4.151E-06 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 1.49E-08 | 1.195E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 1.2E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 2.301E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.67E-07 | 6.699E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 5.959E-07 | 2.048E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 2.617E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 9.999E-07 | 2.673E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 1.648E-06 | 3.964E-04 |
|---|
| GPI anchor biosynthetic process | GO:0006506 |  | 2.526E-06 | 5.526E-04 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.031E-09 | 2.518E-06 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 2.022E-09 | 2.469E-06 |
|---|
| phosphatidic acid metabolic process | GO:0046473 |  | 3.471E-09 | 2.826E-06 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 7.714E-09 | 4.711E-06 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 8.651E-08 | 4.227E-05 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 6.961E-07 | 2.834E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 1.254E-06 | 4.375E-04 |
|---|
| GPI anchor biosynthetic process | GO:0006506 |  | 2.048E-06 | 6.254E-04 |
|---|
| GPI anchor metabolic process | GO:0006505 |  | 2.239E-06 | 6.077E-04 |
|---|
| regulation of smooth muscle cell proliferation | GO:0048660 |  | 2.655E-06 | 6.487E-04 |
|---|
| phosphoinositide biosynthetic process | GO:0046489 |  | 3.636E-06 | 8.075E-04 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.835E-09 | 4.415E-06 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 3.451E-09 | 4.151E-06 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 1.49E-08 | 1.195E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 1.2E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 2.301E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.67E-07 | 6.699E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 5.959E-07 | 2.048E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 2.617E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 9.999E-07 | 2.673E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 1.648E-06 | 3.964E-04 |
|---|
| GPI anchor biosynthetic process | GO:0006506 |  | 2.526E-06 | 5.526E-04 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 8.235E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 5.693E-08 | 1.016E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 6.786E-08 | 8.077E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.495E-07 | 1.335E-04 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.561E-07 | 1.115E-04 |
|---|
| kinase regulator activity | GO:0019207 |  | 3.306E-07 | 1.968E-04 |
|---|
| protein kinase inhibitor activity | GO:0004860 |  | 1.337E-06 | 6.82E-04 |
|---|
| kinase inhibitor activity | GO:0019210 |  | 1.655E-06 | 7.388E-04 |
|---|
| induction of apoptosis by intracellular signals | GO:0008629 |  | 5.001E-06 | 1.984E-03 |
|---|
| phospholipid:diacylglycerol acyltransferase activity | GO:0046027 |  | 6.073E-06 | 2.169E-03 |
|---|
| regulation of retroviral genome replication | GO:0045091 |  | 6.073E-06 | 1.971E-03 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.861E-08 | 6.787E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 4.793E-08 | 8.742E-05 |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 7.557E-08 | 9.189E-05 |
|---|
| mannosyltransferase activity | GO:0000030 |  | 1.206E-07 | 1.1E-04 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.206E-07 | 8.8E-05 |
|---|
| protein kinase regulator activity | GO:0019887 |  | 1.373E-07 | 8.35E-05 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 1.46E-07 | 7.609E-05 |
|---|
| protein serine/threonine kinase inhibitor activity | GO:0030291 |  | 1.854E-07 | 8.454E-05 |
|---|
| kinase regulator activity | GO:0019207 |  | 2.434E-07 | 9.868E-05 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 3.827E-07 | 1.396E-04 |
|---|
| glycerophospholipid metabolic process | GO:0006650 |  | 4.819E-07 | 1.598E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.835E-09 | 4.415E-06 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 3.451E-09 | 4.151E-06 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 1.49E-08 | 1.195E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 1.994E-08 | 1.2E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 4.781E-08 | 2.301E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.67E-07 | 6.699E-05 |
|---|
| protein oligomerization | GO:0051259 |  | 5.959E-07 | 2.048E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 8.7E-07 | 2.617E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 9.999E-07 | 2.673E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 1.648E-06 | 3.964E-04 |
|---|
| GPI anchor biosynthetic process | GO:0006506 |  | 2.526E-06 | 5.526E-04 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.071E-08 | 2.463E-05 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 2.025E-08 | 2.328E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.152E-08 | 3.95E-05 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 5.892E-08 | 3.388E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 9.008E-08 | 4.143E-05 |
|---|
| glycerophospholipid metabolic process | GO:0006650 |  | 1.768E-07 | 6.778E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 2.552E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.076E-06 | 5.968E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 2.898E-06 | 7.406E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.378E-06 | 7.769E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 6.572E-06 | 1.374E-03 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.071E-08 | 2.463E-05 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 2.025E-08 | 2.328E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.152E-08 | 3.95E-05 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 5.892E-08 | 3.388E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 9.008E-08 | 4.143E-05 |
|---|
| glycerophospholipid metabolic process | GO:0006650 |  | 1.768E-07 | 6.778E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 2.552E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.076E-06 | 5.968E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 2.898E-06 | 7.406E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.378E-06 | 7.769E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 6.572E-06 | 1.374E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.071E-08 | 2.463E-05 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 2.025E-08 | 2.328E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.152E-08 | 3.95E-05 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 5.892E-08 | 3.388E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 9.008E-08 | 4.143E-05 |
|---|
| glycerophospholipid metabolic process | GO:0006650 |  | 1.768E-07 | 6.778E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 2.552E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.076E-06 | 5.968E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 2.898E-06 | 7.406E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.378E-06 | 7.769E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 6.572E-06 | 1.374E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.071E-08 | 2.463E-05 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 2.025E-08 | 2.328E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 5.152E-08 | 3.95E-05 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 5.892E-08 | 3.388E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 9.008E-08 | 4.143E-05 |
|---|
| glycerophospholipid metabolic process | GO:0006650 |  | 1.768E-07 | 6.778E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.766E-07 | 2.552E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 2.076E-06 | 5.968E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 2.898E-06 | 7.406E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.378E-06 | 7.769E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 6.572E-06 | 1.374E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| glycerophospholipid biosynthetic process | GO:0046474 |  | 1.035E-08 | 2.456E-05 |
|---|
| glycerolipid biosynthetic process | GO:0045017 |  | 1.913E-08 | 2.269E-05 |
|---|
| phospholipid biosynthetic process | GO:0008654 |  | 5.39E-08 | 4.264E-05 |
|---|
| deoxyribonucleotide biosynthetic process | GO:0009263 |  | 7.782E-08 | 4.616E-05 |
|---|
| nucleoside diphosphate metabolic process | GO:0009132 |  | 1.244E-07 | 5.904E-05 |
|---|
| glycerophospholipid metabolic process | GO:0006650 |  | 2.043E-07 | 8.079E-05 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 7.536E-07 | 2.555E-04 |
|---|
| protein oligomerization | GO:0051259 |  | 1.833E-06 | 5.436E-04 |
|---|
| protein amino acid O-linked glycosylation | GO:0006493 |  | 3.378E-06 | 8.906E-04 |
|---|
| deoxyribonucleotide metabolic process | GO:0009262 |  | 3.378E-06 | 8.015E-04 |
|---|
| positive regulation of smooth muscle cell proliferation | GO:0048661 |  | 7.146E-06 | 1.542E-03 |
|---|



