Study run-a1
Study informations
119 subnetworks in total page | file
406 genes associated page | file
Enriched GO terms page
General informations
General Index page
Study Index page
Subnetwork 10084-0-real
score
| Dataset | Score | P-val 1 | P-val 2 | P-val 3 |
|---|
| Desmedt | 0.1941 | 7.090e-03 | 8.453e-03 | 7.637e-02 |
|---|
| IPC-NIBC-129 | 0.2571 | 7.485e-02 | 9.744e-02 | 5.791e-01 |
|---|
| Ivshina_GPL96-GPL97 | 0.2649 | 4.075e-03 | 6.866e-03 | 1.488e-01 |
|---|
| Loi_GPL570 | 0.2848 | 5.579e-02 | 5.539e-02 | 2.762e-01 |
|---|
| Loi_GPL96-GPL97 | 0.1608 | 2.680e-04 | 1.173e-03 | 7.802e-03 |
|---|
| Parker_GPL1390 | 0.2287 | 1.066e-01 | 1.069e-01 | 5.577e-01 |
|---|
| Parker_GPL887 | 0.0662 | 7.769e-01 | 7.975e-01 | 8.323e-01 |
|---|
| Pawitan_GPL96-GPL97 | 0.3396 | 2.102e-01 | 1.729e-01 | 7.605e-01 |
|---|
| Schmidt | 0.1911 | 7.702e-03 | 1.113e-02 | 1.066e-01 |
|---|
| Sotiriou | 0.3311 | 5.717e-03 | 2.053e-03 | 1.116e-01 |
|---|
| Wang | 0.1745 | 1.094e-01 | 7.636e-02 | 3.873e-01 |
|---|
| Zhang | 0.1974 | 1.133e-01 | 7.608e-02 | 2.683e-01 |
|---|
| Zhou | 0.3629 | 7.441e-02 | 8.012e-02 | 4.439e-01 |
|---|
Expression data for subnetwork 10084-0-real in each dataset
Desmedt |
IPC-NIBC-129 |
Ivshina_GPL96-GPL97 |
Loi_GPL570 |
Loi_GPL96-GPL97 |
Parker_GPL1390 |
Parker_GPL887 |
Pawitan_GPL96-GPL97 |
Schmidt |
Sotiriou |
Wang |
Zhang |
Zhou |
Subnetwork structure for each dataset
- Desmedt
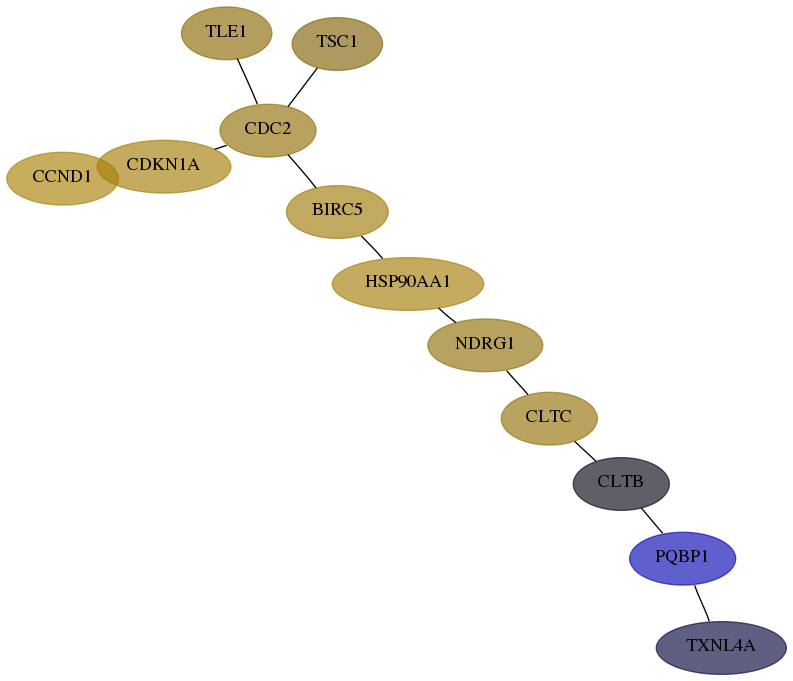
Score for each gene in subnetwork 10084-0-real in each dataset
| Gene Symbol | Links | Frequency | Frequency Rank | Subnetwork score rank | Global rank |
Desmedt | IPC-NIBC-129 | Ivshina_GPL96-GPL97 | Loi_GPL570 | Loi_GPL96-GPL97 | Parker_GPL1390 | Parker_GPL887 | Pawitan_GPL96-GPL97 | Schmidt | Sotiriou | Wang | Zhang | Zhou |
|---|
| cltc |   | 3 | 79 | 330 | 311 | 0.075 | 0.118 | 0.075 | 0.072 | 0.176 | 0.002 | -0.125 | 0.213 | -0.013 | 0.095 | 0.108 | -0.025 | 0.294 |
|---|
| cdkn1a |   | 70 | 3 | 73 | 51 | 0.115 | -0.020 | 0.065 | 0.082 | 0.099 | 0.124 | 0.055 | 0.087 | 0.068 | 0.144 | 0.015 | -0.201 | 0.069 |
|---|
| tle1 |   | 23 | 9 | 78 | 60 | 0.062 | 0.235 | -0.032 | -0.177 | 0.033 | 0.085 | 0.339 | 0.068 | 0.126 | -0.027 | -0.028 | 0.068 | 0.025 |
|---|
| pqbp1 |   | 1 | 164 | 382 | 382 | -0.154 | 0.039 | 0.161 | 0.098 | 0.110 | 0.057 | 0.319 | 0.061 | -0.137 | 0.044 | -0.140 | 0.073 | 0.117 |
|---|
| cltb |   | 3 | 79 | 330 | 311 | -0.002 | 0.119 | 0.171 | 0.109 | 0.083 | 0.160 | -0.334 | 0.074 | 0.056 | 0.073 | 0.089 | -0.007 | 0.223 |
|---|
| hsp90aa1 |   | 11 | 21 | 234 | 204 | 0.116 | 0.030 | 0.164 | 0.076 | 0.119 | 0.155 | -0.199 | 0.169 | 0.069 | 0.102 | 0.052 | -0.011 | 0.399 |
|---|
| ndrg1 |   | 9 | 27 | 250 | 223 | 0.072 | 0.301 | 0.084 | 0.324 | 0.108 | 0.102 | 0.240 | 0.083 | 0.068 | 0.064 | 0.128 | 0.092 | -0.255 |
|---|
| ccnd1 |   | 102 | 2 | 1 | 2 | 0.127 | -0.183 | 0.062 | 0.190 | 0.157 | -0.002 | -0.190 | 0.105 | -0.107 | 0.174 | 0.106 | 0.144 | 0.290 |
|---|
| tsc1 |   | 47 | 5 | 17 | 8 | 0.049 | 0.012 | -0.055 | 0.072 | 0.055 | -0.144 | -0.242 | -0.196 | 0.006 | 0.010 | 0.043 | -0.007 | 0.207 |
|---|
| cdc2 |   | 103 | 1 | 1 | 1 | 0.073 | 0.247 | 0.206 | 0.172 | 0.247 | 0.153 | -0.131 | 0.282 | 0.226 | 0.221 | 0.067 | 0.297 | 0.336 |
|---|
| txnl4a |   | 2 | 104 | 251 | 239 | -0.008 | 0.172 | 0.148 | 0.211 | 0.199 | 0.132 | 0.270 | 0.207 | 0.056 | 0.194 | 0.015 | 0.082 | 0.135 |
|---|
| birc5 |   | 14 | 17 | 162 | 140 | 0.101 | 0.216 | 0.190 | 0.134 | 0.223 | 0.149 | 0.166 | 0.202 | 0.236 | 0.241 | 0.124 | 0.205 | 0.400 |
|---|
GO Enrichment output for subnetwork 10084-0-real in each dataset
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.8E-06 | 0.01104047 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 0.01174456 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 8.579E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.466E-05 | 0.01418207 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.466E-05 | 0.01134566 |
|---|
| protein refolding | GO:0042026 |  | 3.696E-05 | 0.01416801 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.696E-05 | 0.01214401 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.696E-05 | 0.01062601 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01111723 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.991E-05 | 0.01147995 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
IPC-NIBC-129 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.504E-06 | 3.674E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.841E-06 | 2.249E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.839E-06 | 6.384E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.8E-06 | 5.985E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.115E-05 | 5.449E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.175E-05 | 4.785E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.175E-05 | 4.101E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.644E-05 | 5.021E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.644E-05 | 4.463E-03 |
|---|
| response to UV | GO:0009411 |  | 1.684E-05 | 4.115E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.876E-05 | 4.167E-03 |
|---|
Ivshina_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.466E-06 | 5.934E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.122E-06 | 3.755E-03 |
|---|
| interphase | GO:0051325 |  | 3.534E-06 | 2.834E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.092E-05 | 6.566E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.092E-05 | 5.252E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.603E-05 | 6.429E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.636E-05 | 5.624E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.824E-05 | 5.486E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.289E-05 | 6.12E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.289E-05 | 5.508E-03 |
|---|
| response to UV | GO:0009411 |  | 2.462E-05 | 5.385E-03 |
|---|
Loi_GPL570 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.504E-06 | 3.674E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.841E-06 | 2.249E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 7.839E-06 | 6.384E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 9.8E-06 | 5.985E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.115E-05 | 5.449E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.175E-05 | 4.785E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.175E-05 | 4.101E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 1.644E-05 | 5.021E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 1.644E-05 | 4.463E-03 |
|---|
| response to UV | GO:0009411 |  | 1.684E-05 | 4.115E-03 |
|---|
| regulation of cyclin-dependent protein kinase activity | GO:0000079 |  | 1.876E-05 | 4.167E-03 |
|---|
Loi_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.466E-06 | 5.934E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.122E-06 | 3.755E-03 |
|---|
| interphase | GO:0051325 |  | 3.534E-06 | 2.834E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.092E-05 | 6.566E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.092E-05 | 5.252E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.603E-05 | 6.429E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.636E-05 | 5.624E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.824E-05 | 5.486E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.289E-05 | 6.12E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.289E-05 | 5.508E-03 |
|---|
| response to UV | GO:0009411 |  | 2.462E-05 | 5.385E-03 |
|---|
Parker_GPL1390 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 2.306E-08 | 8.235E-05 |
|---|
| clathrin coat of coated pit | GO:0030132 |  | 3.458E-08 | 6.174E-05 |
|---|
| clathrin coat of trans-Golgi network vesicle | GO:0030130 |  | 6.786E-08 | 8.077E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 6.786E-08 | 6.058E-05 |
|---|
| trans-Golgi network transport vesicle membrane | GO:0012510 |  | 9.043E-08 | 6.459E-05 |
|---|
| clathrin vesicle coat | GO:0030125 |  | 2.298E-07 | 1.368E-04 |
|---|
| transport vesicle membrane | GO:0030658 |  | 3.345E-07 | 1.706E-04 |
|---|
| trans-Golgi network transport vesicle | GO:0030140 |  | 5.445E-07 | 2.43E-04 |
|---|
| clathrin coated vesicle membrane | GO:0030665 |  | 1.49E-06 | 5.913E-04 |
|---|
| Golgi-associated vesicle membrane | GO:0030660 |  | 1.832E-06 | 6.541E-04 |
|---|
| vesicle coat | GO:0030120 |  | 2.221E-06 | 7.211E-04 |
|---|
Parker_GPL887 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| cyclin-dependent protein kinase holoenzyme complex | GO:0000307 |  | 1.861E-08 | 6.787E-05 |
|---|
| clathrin coat of coated pit | GO:0030132 |  | 2.79E-08 | 5.088E-05 |
|---|
| clathrin coat of trans-Golgi network vesicle | GO:0030130 |  | 5.475E-08 | 6.657E-05 |
|---|
| trans-Golgi network transport vesicle membrane | GO:0012510 |  | 7.296E-08 | 6.654E-05 |
|---|
| midbody | GO:0030496 |  | 7.296E-08 | 5.323E-05 |
|---|
| cyclin-dependent protein kinase regulator activity | GO:0016538 |  | 1.206E-07 | 7.333E-05 |
|---|
| clathrin vesicle coat | GO:0030125 |  | 1.854E-07 | 9.662E-05 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 1.854E-07 | 8.454E-05 |
|---|
| spindle microtubule | GO:0005876 |  | 2.699E-07 | 1.094E-04 |
|---|
| transport vesicle membrane | GO:0030658 |  | 2.699E-07 | 9.846E-05 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.15E-07 | 1.045E-04 |
|---|
Pawitan_GPL96-GPL97 file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 2.466E-06 | 5.934E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 3.122E-06 | 3.755E-03 |
|---|
| interphase | GO:0051325 |  | 3.534E-06 | 2.834E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 1.092E-05 | 6.566E-03 |
|---|
| activation of Ras GTPase activity | GO:0032856 |  | 1.092E-05 | 5.252E-03 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 1.603E-05 | 6.429E-03 |
|---|
| protein refolding | GO:0042026 |  | 1.636E-05 | 5.624E-03 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 1.824E-05 | 5.486E-03 |
|---|
| positive regulation of mitotic cell cycle | GO:0045931 |  | 2.289E-05 | 6.12E-03 |
|---|
| spindle checkpoint | GO:0031577 |  | 2.289E-05 | 5.508E-03 |
|---|
| response to UV | GO:0009411 |  | 2.462E-05 | 5.385E-03 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.8E-06 | 0.01104047 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 0.01174456 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 8.579E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.466E-05 | 0.01418207 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.466E-05 | 0.01134566 |
|---|
| protein refolding | GO:0042026 |  | 3.696E-05 | 0.01416801 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.696E-05 | 0.01214401 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.696E-05 | 0.01062601 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01111723 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.991E-05 | 0.01147995 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
Sotiriou file
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.8E-06 | 0.01104047 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 0.01174456 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 8.579E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.466E-05 | 0.01418207 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.466E-05 | 0.01134566 |
|---|
| protein refolding | GO:0042026 |  | 3.696E-05 | 0.01416801 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.696E-05 | 0.01214401 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.696E-05 | 0.01062601 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01111723 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.991E-05 | 0.01147995 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.8E-06 | 0.01104047 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 0.01174456 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 8.579E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.466E-05 | 0.01418207 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.466E-05 | 0.01134566 |
|---|
| protein refolding | GO:0042026 |  | 3.696E-05 | 0.01416801 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.696E-05 | 0.01214401 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.696E-05 | 0.01062601 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01111723 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.991E-05 | 0.01147995 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.8E-06 | 0.01104047 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 1.021E-05 | 0.01174456 |
|---|
| interphase | GO:0051325 |  | 1.119E-05 | 8.579E-03 |
|---|
| negative regulation of protein ubiquitination | GO:0031397 |  | 2.466E-05 | 0.01418207 |
|---|
| positive regulation of Rho GTPase activity | GO:0032321 |  | 2.466E-05 | 0.01134566 |
|---|
| protein refolding | GO:0042026 |  | 3.696E-05 | 0.01416801 |
|---|
| spindle checkpoint | GO:0031577 |  | 3.696E-05 | 0.01214401 |
|---|
| establishment of chromosome localization | GO:0051303 |  | 3.696E-05 | 0.01062601 |
|---|
| G1/S transition of mitotic cell cycle | GO:0000082 |  | 4.35E-05 | 0.01111723 |
|---|
| positive regulation of cell cycle | GO:0045787 |  | 4.991E-05 | 0.01147995 |
|---|
| positive regulation of cyclin-dependent protein kinase activity | GO:0045737 |  | 5.169E-05 | 0.01080849 |
|---|
| Name | Accession Number | Link | P-val | Corrected P-val |
|---|
| protein refolding | GO:0042026 |  | 6.175E-08 | 1.465E-04 |
|---|
| outer mitochondrial membrane organization | GO:0007008 |  | 1.08E-07 | 1.281E-04 |
|---|
| chaperone-mediated protein complex assembly | GO:0051131 |  | 1.726E-07 | 1.365E-04 |
|---|
| positive regulation of nitric oxide biosynthetic process | GO:0045429 |  | 3.47E-06 | 2.059E-03 |
|---|
| G2/M transition of mitotic cell cycle | GO:0000086 |  | 4.044E-06 | 1.919E-03 |
|---|
| regulation of nitric oxide biosynthetic process | GO:0045428 |  | 6.136E-06 | 2.427E-03 |
|---|
| response to unfolded protein | GO:0006986 |  | 6.143E-06 | 2.082E-03 |
|---|
| mitochondrial membrane organization | GO:0007006 |  | 6.966E-06 | 2.066E-03 |
|---|
| response to protein stimulus | GO:0051789 |  | 7.823E-06 | 2.063E-03 |
|---|
| interphase of mitotic cell cycle | GO:0051329 |  | 8.582E-06 | 2.037E-03 |
|---|
| interphase | GO:0051325 |  | 9.394E-06 | 2.027E-03 |
|---|



